What We Do
This blog post is our attempt to characterize what we, at NeuroData, do. All of our actions are motivated by our mission, to understand and improve intelligences; the motivation behind our approach is described in our about page. This post summarizes the bulk or our actual work, which can be divided into three complementary threads:
- big data systems
- statistics / machine learning / artificial intelligence
- applications
Big Data Systems
A big data system is a computational ecosystem, including hardware and software, design to support analysis of “big data”, operationally defined as data too big to fit into a single machine’s working memory. The operations of any big data system include: storing, interfacing, pipelining, and visualizing. Existing solutions have been inadequate to support efficient hypothesis generation and scientific discovery. We therefore build, extend, and deploy systems that generalize the functionality of existing systems to support scientific inquiry. We first developed the Open Connectome Project stack in 2011 to host the first ever big dataset in neuroscience, a 10 terabyte electron microscopy dataset from Davi Bock and Clay Read (ocp). As the data size and complexity increased, the development needs extended beyond our capabilities, so we began collaborating extensively with other teams, including scientists and engineers at Google, Allen Institute for Brain Science, and Janelia Research Campus. This collaboration led to our existing open source, community developed, computational ecosystem (ndcloud). The core components of this work include bossDB for big data storage, NDWebtools for interfacing with the data, neuroglancer for visualizing, and various pipelines for different modalities, including ndmg for functional, structural, and diffusion magnetic resonance imaging and reg for registration of 2D and 3D volumes. We continue to extend our collaboration network and ecosystem, to support an ever growing need for big data systems in brain sciences across scales and modalities, ranging from electron microscopy, to whole clear brains, to human and non-human magnetic resonance imaging.
Statistics / Machine Learning / Artificial Intelligence
Statistics, machine learning, and artificial intelligence are terms that describe complementary approaches to solving overlapping sets of problems. Central to all of them is the existence of some data samples, and a question; these approaches then build tools designed to learn an answer to the question from the data. Existing tools, however, have severe limitations that we must overcome in order to obtain answers to the questions of interest. First, raw data in neuroscience tends to be very high-dimensional (e.g., images can be petabytes), and exhibit many nonlinear relationships (e.g., the input/output function of a neuron). We have therefore developed a number computational statistics tools for such settings. This includes state of the art methods for dimensionality reduction (LOL), classification (rerf), time-series modeling [mr. sid]](https://doi.org/10.1016/J.PATREC.2016.12.012), clustering (eclust), and hypothesis testing (mgc). Second, the eventual representation of data most interesting to us is networks in the brain, or connectomes. We have therefore spent much of the last 10 years building foundational statistical estimators, theories, and algorithms for modeling populations of networks with graph, vertex, and edge attributes. A survey summarizing much of our work was recently published in JMLR (rdpg). We subsequently developed a python package that implements all of our theoretical developments (graspy), and wrote a review article on our approach to modeling connectomes, called connectal coding.
Applications
The potential applications of our work are widespread. We illustrate a couple applications spanning our most relevant work, including model (non-human) systems and human variation. First, we characterized which sets of neurons were causally involved in which behaviors, in larval Drosophila. This led to an exploratory analysis of the mushroom body of the larval Drosophila, which we describe in detail in a technical report, with further analysis available in our survey JMLR paper, and our connectal coding paper. Second, to characterize human variation, we developed a cloud pipeline for estimation and analysis of human connectomes (sic), and used it to quantify variability present within and across individuals and studies (ndmg).
Open Science
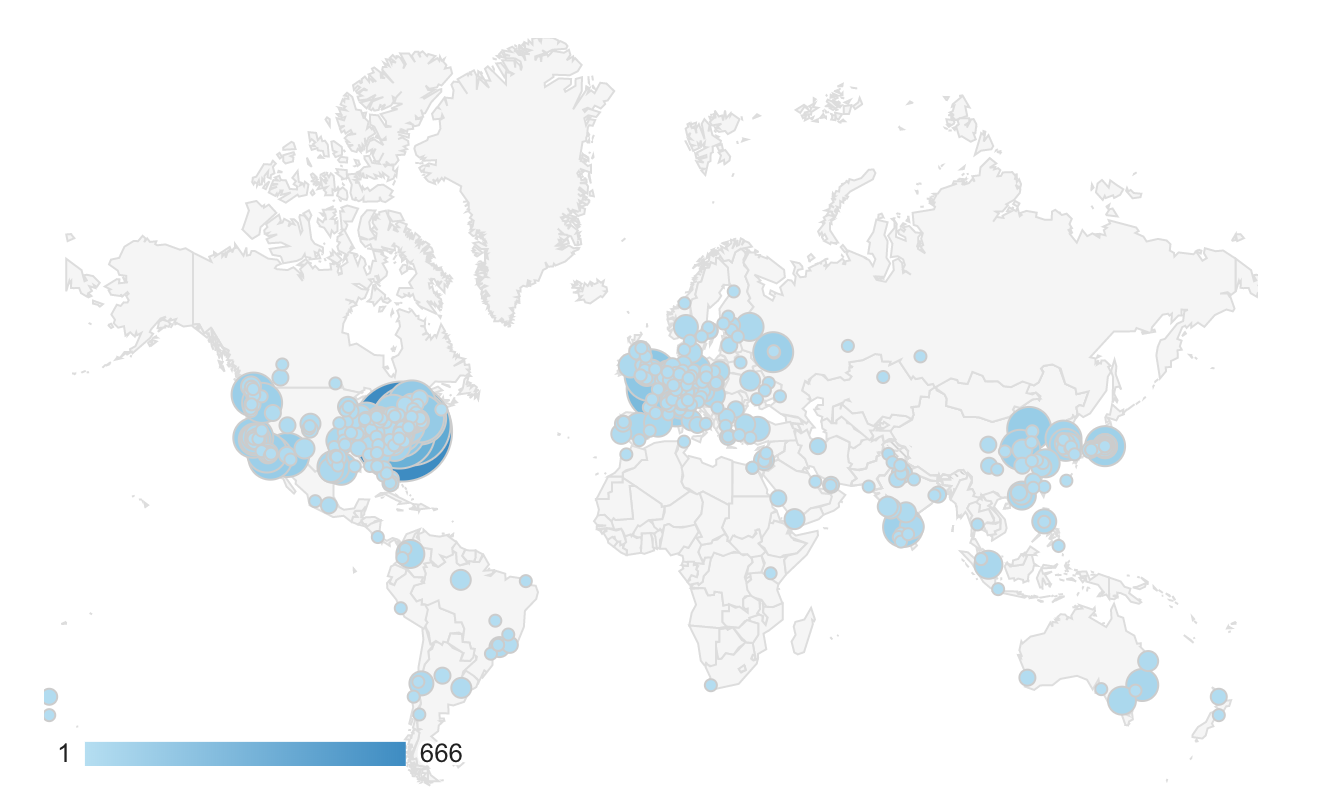
A key principle underlying our work is the democratization of science: we desire that anybody, regardless of resources, is able to both access and contribute to the cutting edge of scientific discovery. To that end, all of the work we do is open science, including both open source code and open access data. Over the last 8 years, our website, https://neurodata.io, has grown from about 100 unique visitors each week to >1,000 unique visitors each month. In the last calendar year, >13,000 unique visitors browsed the site. These visitors span every inhabited continent, including over 2,000 different cities (see image below). Over the 8 years, approximately 80,000 unique individuals have visited our site, over double the number of people at a typical Society for Neuroscience conference, suggesting that many non-neuroscientists have visited our site. We hope that those people visiting the site feel the same sense of awe and inspiration as we do, associated with unraveling the secrets of mental function in these beautiful images of brains.